
Chromosome architecture and dynamics
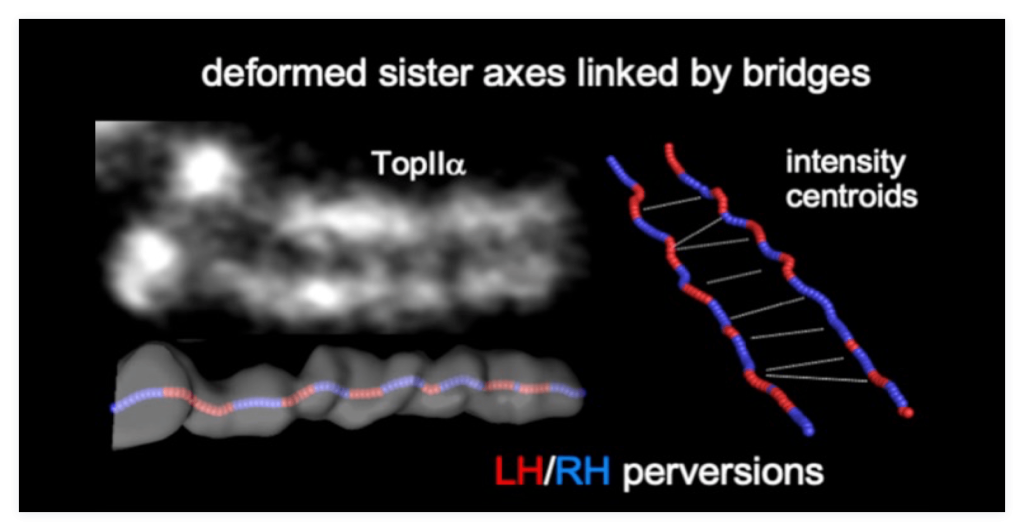
The precise assembly of chromosomes is the basis for maintaining cell cycle stability. Current research on chromosomes mainly focuses on the structure and chemical regulation of components, but there is still a lack of in-depth understanding of its dynamic assembly process and internal driving forces. Our previous studies suggested that chromosome assembly is mediated by its internal mechanical stress, and internal mechanical stress promotes sister resolution at late prophase, leading to evenly spaced inter-sister bridges; it also promotes sisters’ global parallel separation at anaphase, upon cohesin cleaved by separase. We also suggested chromosomes comprise co-oriented linear loop/axis arrays that linearly shorten, and chromosome is not helically coiled, but organized in sequential half-helices of alternating handedness. Those work opened a new insight for chromosome biology research.
Next, we plan to study the molecular and physical mechanism of chromosome mechanical stress, and its dynamics and implication in cell cycle.
Chromosome instability and innate immunity
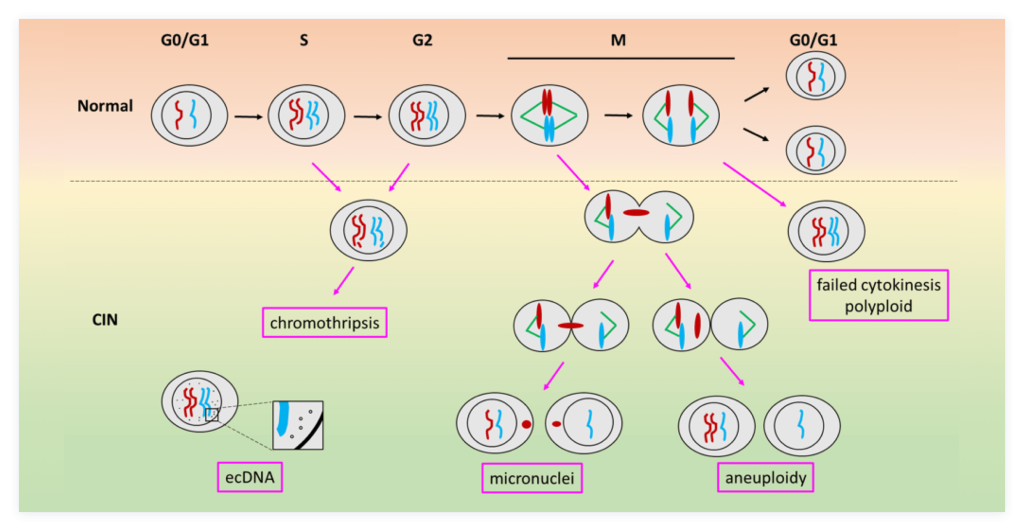
Chromosomal instability is considered an important feature of cancer cells. Abnormal chromosome replication, organization, and segregation can lead to chromosomal instability, resulting in aneuploidy, micronuclei, chromosome fragmentation, or ecDNA, etc. Take ecDNA for example, it has high levels in cancer cells. However, the mechanism by which it arises remains unknown.
To study the mechanisms of chromosome instability, we are building a chromosome instability platform and trying to find some key molecules that are involved in those progress. We will examine their contribution to the Sting-cGAS pathway, an important sensor of exogenous gene invasion or abnormal self-gene release. This project will provide cytology basis for diagnosis and treatment of chromosomal instability diseases.
Cell dynamics and AI
Cells are the smallest unit of life activities, and qualitative and quantitative research on cell life activities can predict personal health status. In order to qualitatively and quantitatively capture the dynamic occurrence and development of fine tissue structures at the cellular level and subcellular level, we conduct high-temporal-spatial resolution microscopic tracking imaging of the dynamics of cells and subcellular structures, and conduct in-depth artificial intelligence analysis through image recognition and machine learning to capture the rules of cell life activities under normal conditions and provide individuals with health predictions at the cellular level .